Editor’s note: This is the first post in a series titled, “Scalable and Dynamic Data Pipelines.” This series will detail how we at Maxar have integrated open-source software to create an efficient and scalable pipeline to quickly process extremely large datasets to enable users to ask and answer complex questions at scale.
Maxar has a passion to create and deliver Earth Intelligence solutions that enable our customers to discover insights when and where it matters. I am a software engineer on a larger team supporting a mission-driven customer, and we are routinely asked to help analysts make sense of new, unique and incredibly large datasets that come in a variety of formats, sizes and complexities. Typically, we focus on processing large, non-imagery based, geo-temporal data sets at a speed that can quickly evaluate and expose insights critical to mission objectives. My team developed a robust data pipeline that allows us to rapidly integrate new datasets into our petabyte-scale data store. In the first part of this blog series titled, “Scalable and Dynamic Data Pipelines,” we will provide an overview on how we’ve integrated open-source software to create an efficient and scalable Extract, Transform and Load (ETL) process to meet user requirements.
ETL Components
By establishing a standard ETL process, we can routinely collect data, convert it from one form to another, and write the data into a database or data model. Within this process, we use the following tools:

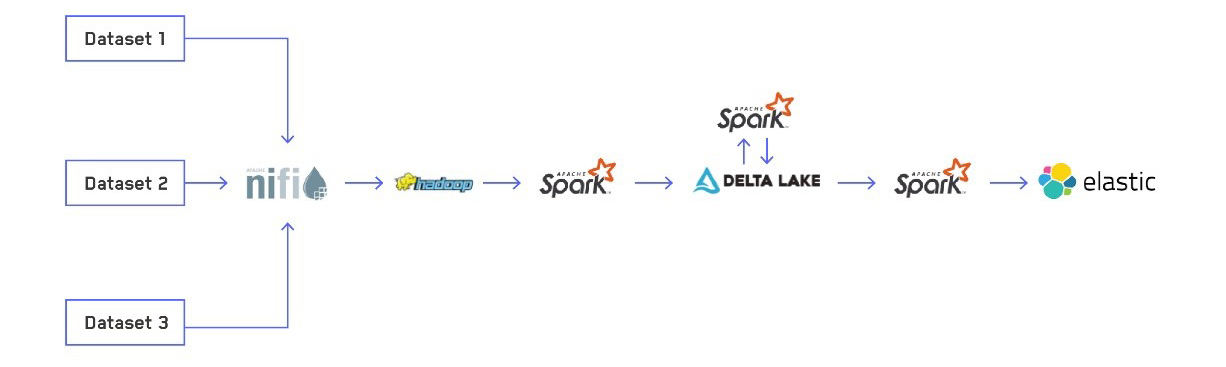
Overview of the ETL pipeline
Our ETL process has four steps:

Benefits to this Approach
- Data exposure: Users can access our data in two ways:
- Elasticsearch for fast query and retrieval
- Delta Lake tables for custom batch analytics
- Application Integration: Our custom applications and visualizations query Elasticsearch as users interact with and explore the data in real time.
- Custom Analytic Support: Users can access our Delta Lake tables in HDFS directly through JupyterHub and use our bare-metal Yarn cluster to run large-scale jobs.
- Discoverability: We register our Delta Lake tables in our Hive Metastore thanks to the recent Delta Lake OSS integration with the new Spark 3 Catalog API, which allows users to easily discover available datasets through the Spark catalog.
- Accessibility: Access to restricted datasets can be controlled by file permissions on our Kerberized Apache Hadoop cluster.
Let's dive into each step in more detail:
STEP 1 - Receiving data via NiFi
Why we use NiFi:
Apache NiFi provides a user-friendly interface for collecting data from various sources and depositing it in a centralized location. NiFi allows both pulling from external APIs and setting up remote input and output ports, creating an easy way to share data with our partners.
How we use it:
Maxar uses NiFi to move data and place it in the correct location in HDFS. Transformations are limited to dealing with functions Spark is not suited for, such as extracting .zip or .tar archives. We partition our raw data by day for most of our datasets. If a feed has a date embedded in the file name, we use that date as the partition folder to place the file into. If it does not have a date in the file name, we place it into a partition for the current day. Because we use Spark structured streaming for our ETL process, we don't need to worry about data arriving late or being inserted into an old partition.
Lessons learned:
In the past we attempted doing our ETL process via NiFi, but it became incredibly hard to maintain. A drag-and-drop interface seems simple to use at first, but once your pipeline becomes 100 processors long, it's a nightmare to try to make updates or debug errors. Additionally, we had issues scaling our NiFi cluster. Several times we had to restart one or all our NiFi nodes because the cluster state become out of sync, and load balancing queues occasionally freeze for unknown reasons. We recently updated our NiFi cluster from version 1.9.2 to 1.11.4, and we are hopeful some of these issues resolve themselves. NiFi also quickly hits IO bottlenecks as each transformation requires rewriting data to disk.
STEP 2 - Processing the raw data into Delta Lake tables
Why we use Spark Streaming and Delta Lake:
Apache Spark structured streaming provides robust, fault-tolerant, distributed processing at scale. Each source file is tracked individually, so there is no need for batch processing entire partitions at a time or worrying about late-arriving data. Combining our Spark processing with the Delta Lake table format enables consistent and efficient table metadata handling with exactly-once guarantees and allows us to unlock the full potential of our hardware.
How we use it:
Once the data is written to HDFS in its raw state, a Spark structured streaming application picks it up for transformation and writes it to a Delta Lake table. We built a custom library that abstracts out the logic for building production-ready Spark applications, as well as common transformations we perform on all our datasets. We try to limit the unique logic per dataset as much as possible, which is usually limited to figuring out how to load the raw data into a Spark DataFrame and determining the import column names we want to enrich. From that point, the library can take over the rest of the processing, applying common normalizations and enrichments, and writing the output to the appropriate Delta Lake table.
The basic steps for creating a pipeline for a new dataset include:
- Manually interact with the data in a PySpark session, deriving a schema and determining the fields it contains and the transformations we want to apply
- Build the pipeline using our processing library and test out in batch mode
- Once the pipeline is ready, start the application in streaming mode
Lessons learned:
Deriving the schema and determining the necessary transformations is the most time-consuming task as it takes the greatest customization per source. Having data in a common Delta Lake format makes downstream processes much easier to develop. In Part 2 of this blog series, we will cover more details of our processing library as well as common hurdles we've faced incorporating new datasets.
STEP 3 - Optimizing Delta Lake tables
Why Optimize Delta Lake tables
Streaming pipelines typically result in many small files, which are highly inefficient for large-scale analysis. Delta Lake allows us to optimize our file sizes after the fact, allowing for both low-latency feeds to downstream systems as well as optimized query performance for batch analytics.
How we use it:
Once per day we run a batch job that goes through each partition in our Delta Lake tables and rewrites the whole partition to a more optimized and consistent set of file sizes, if necessary. In Part 3 of this blog series, we will go into a deep dive of how this process works.
Lessons learned:
The so-called small files problem is a well-known issue in Hadoop. Before we began optimizing our file sizes, we started to run into issues with our NameNode scaling to the number of files we had in our HDFS cluster. The NameNode requires a certain amount of memory per file, so the total number of files you can have in your cluster is proportional to the amount of heap your NameNode has. Optimizing our file sizes had the additional benefit of reducing the total number of files in our cluster, killing two birds with one stone.
STEP 4 - Elasticsearch indexing
Why Elasticsearch
Elasticsearch provides incredibly fast and versatile querying capabilities, allows users to explore new datasets in real time, and scales to the petabyte-size data with which we work.
How we use it:
The final step in the process is indexing the data into Elasticsearch. We use the Elasticsearch-Hadoop library's Spark integration to stream data from our Delta Lake tables to our Elasticsearch cluster. We've gone through several iterations of Elasticsearch configurations and bulk index settings to find ways to index data on the petabyte scale. Our custom applications use Elasticsearch’s expansive query language to allow our end users to explore, analyze and visualize incredibly large datasets in real time.
Lessons learned:
Our hardware contains both hard-disk drives (HDD) and non-volatile memory express drives (NVMe). We initially ran separate Elasticsearch clusters on our HDD and NVMe, putting any dataset small enough on the NVMe and larger datasets on the HDD. Indexing at scale onto spinning disks presents several challenges, and we found methods to greatly increase our throughput. However, we eventually reconfigured to run a single hybrid Elasticsearch cluster using node attributes to run a hot/warm architecture. With this setup we can index all datasets to the NVMe and move older data in time-based indices to the HDD for historical queries. Part 4 of this blog series will provide more information on how we initially tuned our indexing for HDD and how we migrated to a hot/warm architecture that has greatly simplified our workload.
Conclusion
This is the high-level flow of how we transform datasets to make usable by our customers. In further entries of this series, we will dig into the details of each step in this process. Part 2 will explore our process for loading the raw data, transforming it and saving it to our data warehouse for discoverability and efficient batch analytics.
Apache, the Apache feather logo, Apache Hadoop® , Apache Spark™, Apache NiFi™ and the corresponding project logos are trademarks of The Apache Software Foundation. Delta Lake and the Delta Lake logo are trademarks of LF Projects, LLC.